
+91-44-42271500
vrf@snmail.org

+91-44-42271500
vrf@snmail.org

Head of the Department/ chief scientist & senior professor
ONGC Genetics and Molecular Biology Laboratory
Dr. S. Mathavan is currently a chief scientist & senior professor, Head of the Department of SN ONGC department of genetics and molecular biology, Vision Research Foundation, Chennai. Previously,he was working as professor and Head of the Department of genetics at Madurai Kamaraj University, Madurai, Head of zebrafish research group at the Genome Institute of Singapore, Singapore and Genetics and Genomic specialist at LKC School of medicine, Nanyang Technological University, Singjapore. Dr. Mathavan served as overseas visiting senior scientist at University of California, Davis, USA; University of Lyon, France and University of Oslo, Norway, and National University of Singapore, Singapore. Dr.Mathavan has operated multiple international collaborations thus, indicating his leadership ability to operate international collaborations. Some of the overseas collaborations were between University of Oslo, Norway; University of Sheffield, UK and University of Otego, New Zealand. These projects were funded by Government of India (during my tenure in India) and by Government of Singapore, Government of New Zealand (during his tenure in Singapore).
So far 25 students have completed Ph.D. degree under his supervision and he has also trained postdoctoral fellows. Students trained in his lab are placed in leading research institutions all around the world. He has published more than 100 research papers in international peer reviewed journals. Recently, his research has been funded by DBT, New Delhi under Indo-US collaboration to study genetic architecture of diabetic retinopathy.
During his tenure in India (MK University, Madurai), he was the first to clone growth hormone gene from Indian food fish and to generate transgenic food fish which exhibited faster and bigger growth. Subsequently, during his tenure in Singapore, he served as a fulcrum for international collaborations on the genomics projects using fish as developmental and disease model system. His laboratory discovered the process of cytoplasmic polyadenylation in a cohort of maternal RNA and showed that this phenomenon regulated the process of maternal to zygotic transition. He has extensively used microarray and RNA sequencing technologies and has expertise in transcriptome analysis and in discovery of novel mRNA and miRNA transcripts.
After his relocation to Vision Research Foundation, Dr.Mathavan is employing genomics tools and technologies for ophthalmological research. His group is interested in ‘molecular genomics’ to discover transcriptome (coding and long non-coding RNA and microRNAs)in vasculopathies and are employing NGS technology for deep sequencing to explore dynamics of transcriptomics in patient blood samples and retinal cell lines. The long term goal of his lab is to discover RNA biomarkers for vasculopathies and to develop a miRNA biosensors.
Diabetes and its complications are leading cause of DR in Indian T2DM cohorts. The current challenge of diagnosing and identifying the disease in the early stage can prevent blindness. This study will help develop AI algorithms which can be used for diagnosing and predicting the risk of progression of the disease. Using the big data of EMR and retinal imaging [fundus photography and Optical Coherence Tomography (OCT)] at Sankara Nethralaya, Chennai and Madras Diabetes Research Foundation, Chennai, Vellore institute of technology, Chennai along with international technology partners would be able to develop a deep learning algorithm which would be able to do the following
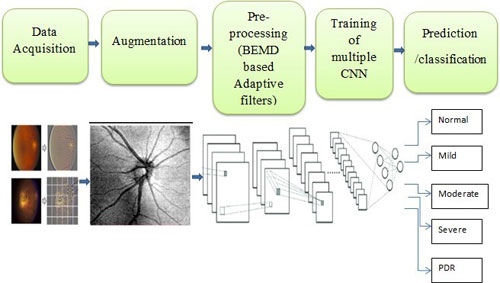
The Genetics of Diabetic Retinopathy (DR) is one of the major projects in Dr.Mathavan’s laboratory which is funded by DBT, New Delhi under an Indo-US collaborative program. Dr. Rajiv Raman, and Dr. S. Sripriya, VRF are also the investigators. The US collaborators are Dr. Sudha Iyengar from Case Reserve Western University and Dr. Lucia Sobrin from The Massachusetts Ear and Eye Infirmary. We are focusing on identifying common and rare variants in the South Indian population using Whole Genome Sequencing (WGS) and Genome-Wide Association Studies (GWAS).
DR is a microvascular complication caused by high blood sugar. It is the main cause of vision loss in the working-age population worldwide. Some of the major risk factors for retinopathy are prolonged duration of diabetes, poor glycemic control, and dyslipidemia. It has been observed that retinopathy is not only influenced by environmental and lifestyle factors but, genetics and epigenetics also play a major role in its’ risk and progress. Thus, it is important to investigate and understand the genetics of this disease.
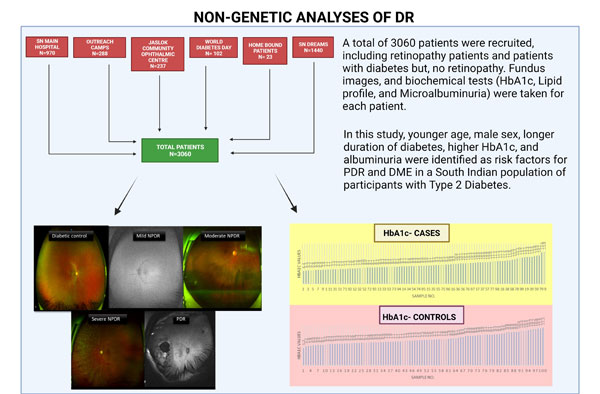
1. Non-genetic analysis of DR:
A total of 3060 patients were recruited by the group for this project. OCT-A and fundus images were taken for each patient. The images were used to grade the patients for their DR stage using the International DR Severity scale. Biochemical tests (HbA1c, Lipid Profile, and Microalbuminuria) were done for the patients. These values were correlated with the image grading for each patient. In this study, younger age, male sex, longer duration of diabetes, higher HbA1c, and albuminuria were identified as risk factors for PDR and DME in a South Indian population of participants with Type 2 Diabetes.
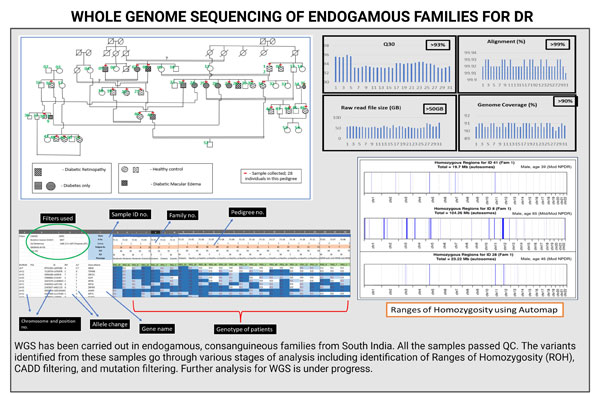
2. WGS:
For WGS study, DR, diabetic, and healthy patients were recruited from large endogamous families of South Indian origin. A total of 32 patients were recruited and whole genomes of these patients were sequenced. All the samples passed QC. Common and rare variants will be identified for DR from the generated genome data. The analysis is in progress for this data.
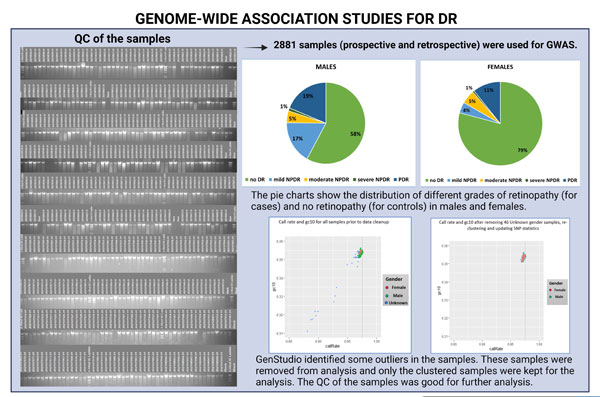
3. GWAS:
The GWAS data has been generated and analysis is in progress jointly with the US collaborators. A total of 2881 samples were subjected to GWAS study. Some samples were identified as outliers and these samples were removed. Only the clustered samples were used for further analysis. All the remaining samples passed QC and the analysis is in progress for this data. We aim to identify rare variants that will be important to the development and progress of DR. The identified variants can be used in the future as biomarkers to identify retinopathy as well as using them as therapeutic agents.
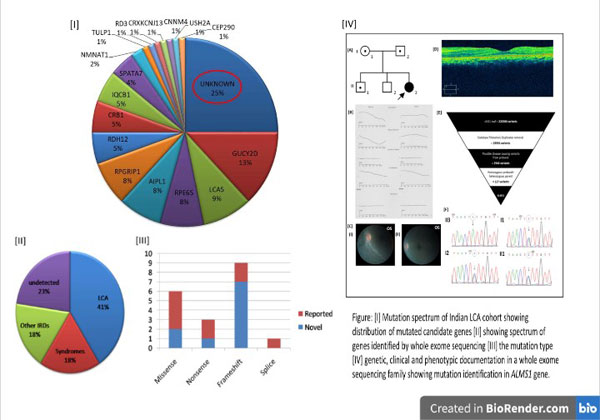
Identification of Novel mutations in Indian LCA cohorts:
The mutation prevalence in known candidate genes in the Indian cohort remained unknown until we published our data on targeted resequencing in a large Indian LCA cohort of 92 cases. The data showed 75% to be mutation positive in known candidate genes (Figure I). The above project gave us hope of screening the rest of the mutation negative cases by whole exome sequencing, anticipating the identification of one or more novel gene(s) in these set of LCA cases. Whole exome sequencing and analyses further identified the disease causing pathogenic variants in 77% of the mutation negative cases (Figure II). Our cohort reports 52% novel variants not detected in other population. These identified variants were 47% frameshifts, 32% missense, 15% nonsense and 5% splice variants (Figure III). Upon molecular diagnosis in the current study, reanalysis of the phenotypes of the patients helped in rediagnosis of 36% cases as either syndromic or nonsyndromic IRDs (Figure IV).
The study so far had helped us in identifying one novel candidate gene for LCA and had opened up an avenue for whole genome sequencing analysis to detect disease causing deep intronic variants or novel genes in the remaining negative cases
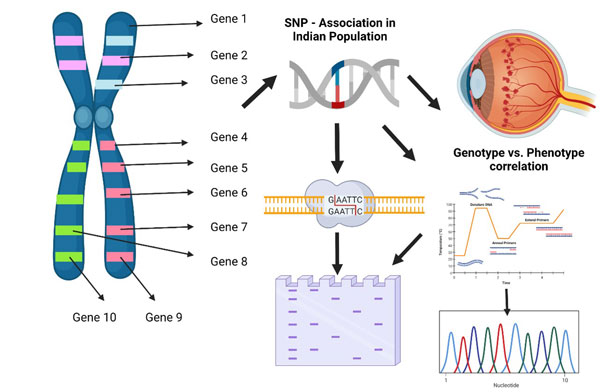
Genotype and Phenotype correlation in ocular vasculopathy
Single nucleotide polymorphism is the most common genetic variants among the wide population. It is one of the well known biomarker, supporting the research to identify and located the disease associated variations. The development of SNP markers allows to automatize and enhance ten folds the effectiveness of genotype analysis to further correlate with phenotype of the disease. In our study, we have chosen, total of 16 SNP highly correlated with the disease from different population. The risk of disease occurrence is correlated with each SNP in Indian population as a prime attempt. To genotype the samples we used various techniques as follows, Allele specific polymerase chain reaction (AS-PCR), Restriction enzyme, Sanger sequencing. The optimized Statiscal significance were performed. Phenotype of the disease is been correlated with the genotype to understand the disease risk and severe. This study will support clinicians for precise disease diagnosis and prognosis.
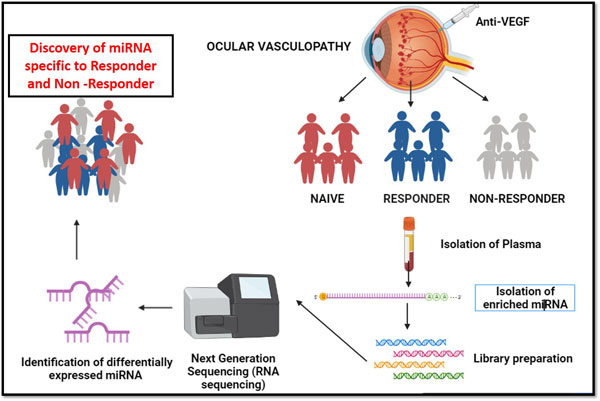
MicroRNA biomarker for anti- VEGF responder and non responder in ocular vasculopathy
MicroRNA plays a important role as biomarker in the field of pharmacogenomics to identify the drug treatment effectives in many diseases like oncology, tuberculosis etc. In ocular vasculopathy, we attempted to identify and discover the anti–VEGF drug responsive and non-responsive biomarker to serve patients to opt for different treatment options, since each anti- VEG injection cost around 35,000 INR approximately. We collected blood samples for anti- VEGF diabetic control, naïve, responder and non – responders. Small RNA were extracted from all the collected samples. Next generation small RNA sequencing was performed for anti-VEGF responders, non-responders and naïve ocular vasculopathy patient samples. The outcome of the project revealed that miRNA-142 is the biomarker for naïve and miRNA-664a is the biomarker for anti-VEGF non-responder patients. Further work is under progress to miRNA biomarkers for DME patients
Google Scholar : Check out this link
1998-1999
UGC Research Award, University Grants Commission, New Delhi, India
1995
Seth Baldeodas Award,Government of India, New Delhi, India
1998
Fellowship: Fellow of National Academy of Sciences, India (FNASc)
Chairman - M.Sc. Biology, Madurai Kamaraj University, Madurai1994 - 1997
Member - Board of Studies, Madurai Kamaraj University, Madurai 1997 - 2000
Member - Board of Studies,Madurai Kamaraj University, Madurai 1992 - 1995
Member - Board of Studies, Mysore University, Mysore 1996 - 1999
Member - Board of Studies, Fatima College, Madurai
Member - Board of Studies, Lady Doak College, Madurai
Member - Board of Studies, ANJA College, Sivakasi
Dr.Sudha K. IyengarSudhaIyangar, Case Western Reserve University, Cleveland, Ohio 44106, USA. Project : Unravelling the genetic architecture of diabetic retinopathy in South India
Dr. Peter Alestrom, (University Oslo, Oslo , Norway ). Project :Zebrafish early embryonic transcriptome and embryonic stem cells.
Dr. PhillipeCollas (University of Oslo, Olso, Norway). Project: Epigenetic and pre-patterning during maternal-zygotic transition (MZT) in zebrafish.
Dr. Julia Horsfield (University of Dunedin, Dunedin, New Zealand). Project: Epigenetic profiling and DNA methylaiton analysis during liver tumor induction and disease progress , using zebrafish model.
Dr. Freek van Eden (University of Sheffield, Sheffield, UK). Project: Sheffield-ASTAR collaboration- Genome-wide binding site analysis of Hif1a inzebrafish.
Dr. Sukla Gosh (University of Calcutta, Calcutta, India). Project: Zebrafish nerve card regeneration
Dr. Gong (University of Singapore, Singapore). Project: Liver tumor analysis using zebrafish model
Prof.Phil Ingham (ASTAR, IMCB, Singapore). Zebrafish model to predict organ toxicities
Molecular Genomics: Discovery of microRNA in Zebrafish and generation of microRNA microarray platform for zebrafish.
Agilent and Molecular Genomics: Exon array for zebrafish. Array design, development and validation.
Sciencewerk : MicroRNA and their role in early embryonic development
Served as Visiting Professor in Bharathidasan University, Trichirapalli, India.
Served as Overseas Consultant to University of Calcutta for a project funded by DST,Government of India.
Served as Adjunct Professor (Honorary) in SRM University, Chennai
Served as Task Force Member: Department of Biotechnology, Aquaculture Taskforce.

Ms Lakshmi priyanka
Research Student

Ms Rizza
Research Student

Mr Murugapandian
Social Worker

Ms Rehana
Optomerician

Ms Lakshmi priyanka
Research Student

Ms Rizza
Research Student

Mr Murugapandian
Social Worker

Ms Rehana
Optomerician
Vision Research Foundation
No. 41 (old 18), College Road,
Chennai - 600 006, Tamil Nadu , India.
Ph No: +91-44-42271500,
+91-44-2827 1616,
vrf@snmail.org
